Gradient Boosting Machines (GBMs)
- GBMs are extremely popular, successful across many domains and one of the leading methods for winning Kaggle competitions.
- GBMs build an ensemble of flat and weak successive trees with each tree learning and improving on the previous.
-
When combined, these trees produce a powerful “committee” often hard to beat with other algorithms.
- The following slides are based on UC Business Analytics R Programming Guide on GBM regression
The idea of GBMs
- Many machine learning models are founded on a single predictive model (i.e. linear regression, penalized models, naive bayes, svm).
- Other approaches (bagging, random forests) are built on the idea of building an ensemble of models where each individual model predicts the outcome and the ensemble simply averages the predicted values. <!–
- The family of boosting methods is based on a different, constructive strategy of ensemble formation. –>
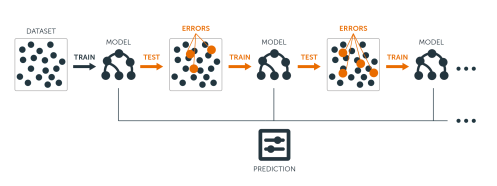
- The idea of boosting is to add models to the ensemble sequentially.
- At each particular iteration, a new weak, base-learner model is trained with respect to the error of the whole ensemble learnt so far.
 { height=70% }
{ height=70% }
Advantages of GBMs
Predictive accuracy
- GBMs often provide predictive accuracy that cannot be beat.
Flexibility
- Optimization on various loss functions possible and several hyperparameter tuning options.
No data pre-processing required
- Often works great with categorical and numerical values as is.
Handles missing data
- Imputation not required.
Disadvantages of GBMs
GBMs overemphasize outliers
- This causes overfitting.
- GBMs will continue improving to minimize all errors. Use cross-validation to neutralize.
Computationally expensive
- GBMs often require many trees (>1000) which can be time and memory exhaustive.
- The high flexibility results in many parameters that interact and influence heavily the behavior of the approach (number of iterations, tree depth, regularization parameters, etc.).
- This requires a large grid search during tuning.
Interpretability
- GBMs are less interpretable, but this is easily addressed with various tools (variable importance, partial dependence plots, LIME, etc.).
Important concepts
Base-learning models
- Boosting is a framework that iteratively improves any weak learning model.
- Many gradient boosting applications allow you to “plug in” various classes of weak learners at your disposal.
- In practice, boosted algorithms often use decision trees as the base-learner. <!–
- Consequently, this tutorial will discuss boosting in the context of regression trees. –>
Training weak models
- A weak model has an error rate only slightly better than random guessing.
- The idea behind boosting is that each sequential model builds a simple weak model to slightly improve the remaining errors.
- Shallow trees represent weak learner - trees with only 1-6 splits.
Benefits of combining many weak models:
- Speed: Constructing weak models is computationally cheap.
- Accuracy improvement: Weak models allow the algorithm to learn slowly; making minor adjustments in new areas where it does not perform well. In general, statistical approaches that learn slowly tend to perform well.
- Avoids overfitting: Making only small incremental improvements with each model in the ensemble allows us to stop the learning process as soon as overfitting has been detected (typically by using cross-validation).
Sequential training with respect to errors
- Boosted trees are grown sequentially;
- Each tree is grown using information from previously grown trees.
- $x \rightarrow$ features and $y \rightarrow$ response:
- The basic algorithm for boosted regression trees can be generalized:
1.) Fit a decission tree: $F_1(x)=y$
2.) the next decission tree is fixed to the residuals of the previous: $h_1(x)=y-F_1(x)$
3.) Add this new tree to our algorithm: $F_2(x)=F_1(x)+h_1(x)$
4.) The next decission tree is fixed to the residuals of $h_2(x)=y-F_2(x)$
5.) Add the new tree to the algorithm: $F_3(x)=F_2(x) + h_1(x)$
Continue this process until some mechanism (i.e. cross validation) tells us to stop.
Boosted regression decision stumps as 0-1024 successive trees are added.
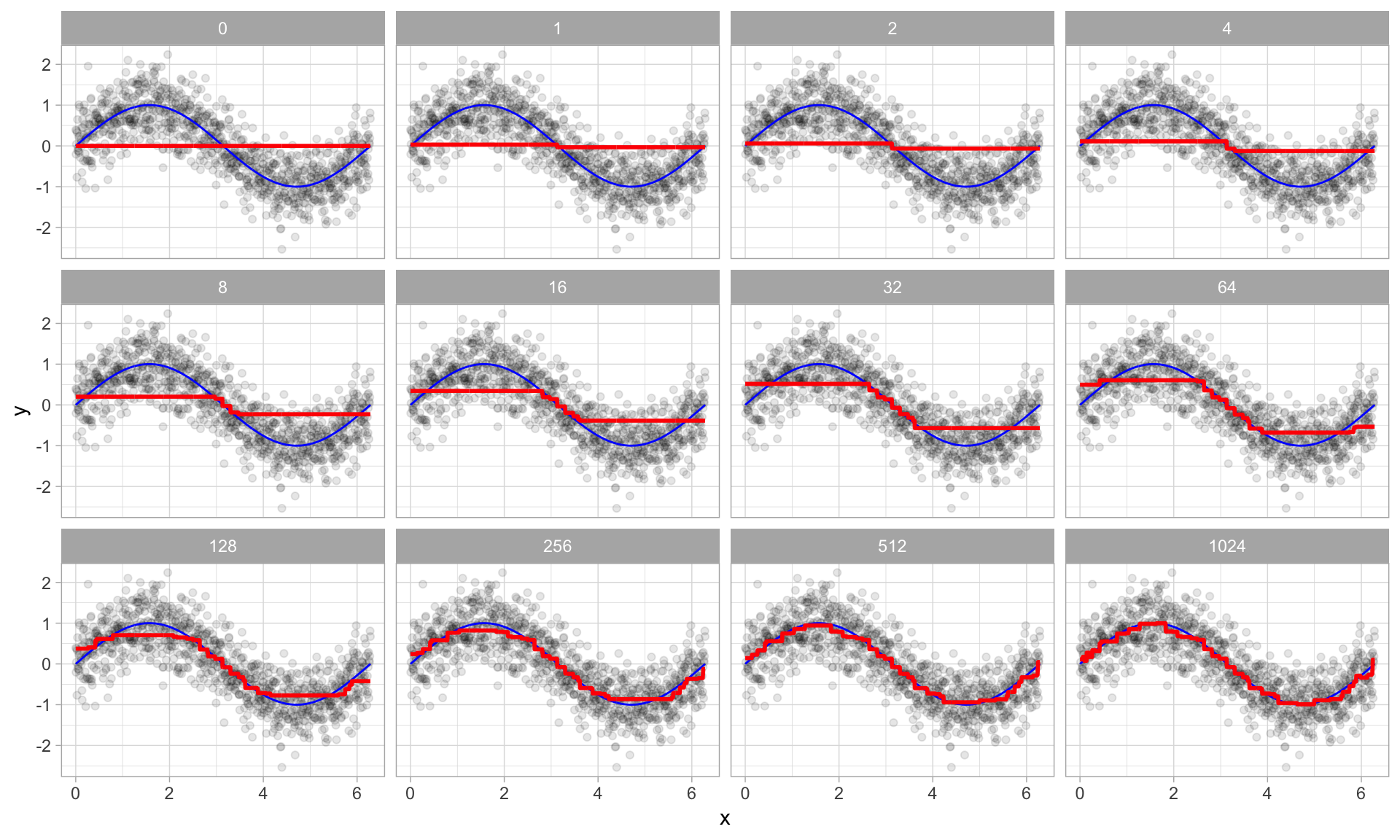
Boosted regression figure - explained
- The figure illustrates a single predictor ($x$) that has a true underlying sine wave relationship (blue line) with y along with some irriducible error.
- The first tree fit in the series is a single decision stump (i.e., a tree with a single split).
- Each following successive decision stump is fit to the previous one’s residuals.
- Initially there are large errors, but each additional decision stump in the sequence makes a small improvement in different areas across the feature space where errors still remain.
Loss functions
- Many algorithms, including decision trees, focus on minimizing the residuals and emphasize the MSE loss function.
- In GBM approach, regression trees are fitted sequentially to minimize the errors. <!–
- This minimizes the loss function - mean squared error (MSE). –>
- Often we wish to focus on other loss functions such as mean absolute error (MAE)
- Or we want to apply the method to a classification problem with a loss function such as deviance.
- With gradient boosting machines we can generalize the procedure to loss functions other than MSE. <!–
- The name gradient boosting machines come from the fact that this procedure can be generalized to loss functions other than MSE. –>
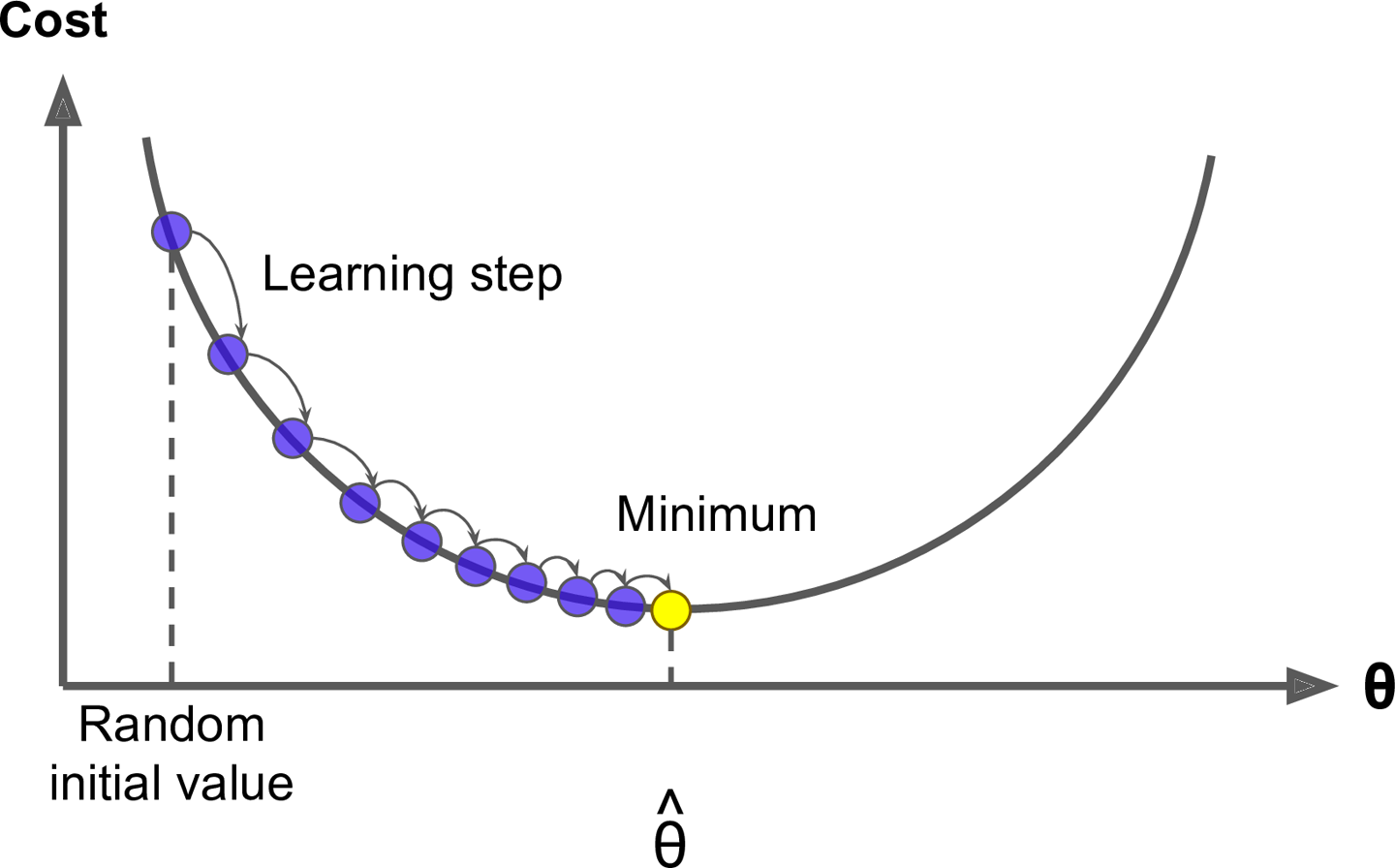
A gradient descent algorithm
- Gradient boosting is considered a gradient descent algorithm.
- Which is a very generic optimization algorithm capable of finding optimal solutions to a wide range of problems.
- The general idea of gradient descent is to tweak parameters iteratively in order to minimize a cost function.
Example
- Suppose you are a downhill skier racing against your friend.
-
A good strategy to beat your friend is to take the path with the steepest slope.
- This is exactly what gradient descent does - it measures the local gradient of the loss (cost) function for a given set of parameters ($\Phi$) and takes steps in the direction of the descending gradient.
- Once the gradient is zero, we have reached the minimum.
Gradient descent (Geron, 2017).
Gradient descent
- Gradient descent can be performed on any loss function that is differentiable.
- This allows GBMs to optimize different loss functions as desired
- An important parameter in gradient descent is the size of the steps which is determined by the learning rate.
- If the learning rate is too small, then the algorithm will take many iterations to find the minimum.
- But if the learning rate is too high, you might jump cross the minimum and end up further away than when you started.
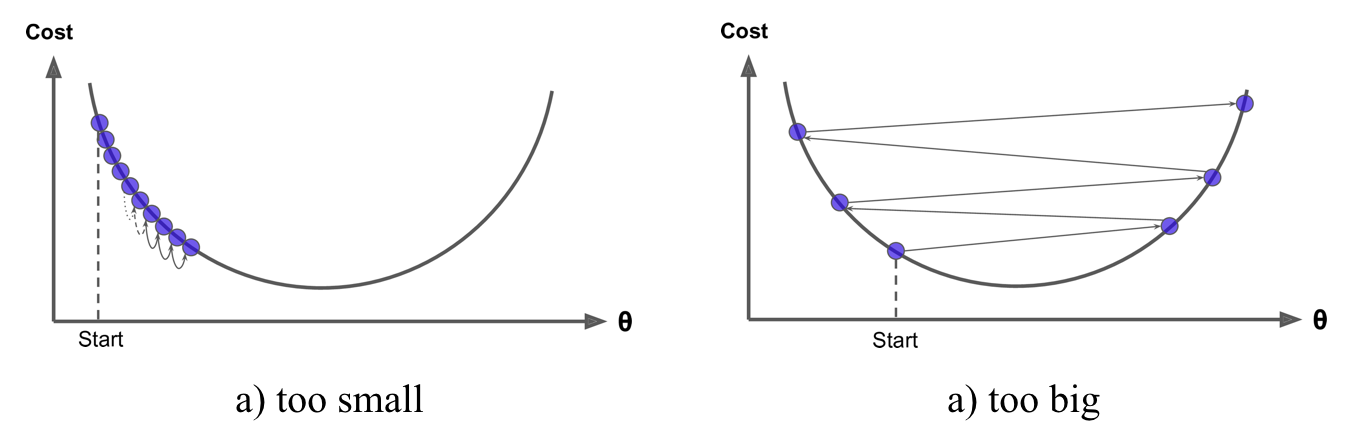
Shape of cost functions
- Not all cost functions are convex (bowl shaped).
- There may be local minimas, plateaus, and other irregular terrain of the loss function that makes finding the global minimum difficult.
- Stochastic gradient descent can help us address this problem.
- Stochastic because the method uses randomly selected (or shuffled) samples to evaluate the gradients.
- By sampling a fraction of the training observations (typically without replacement) and growing the next tree using that subsample.
- This makes the algorithm faster but the stochastic nature of random sampling also adds some random nature in descending the loss function gradient.
- Although this randomness does not allow the algorithm to find the absolute global minimum, it can actually help the algorithm jump out of local minima and off plateaus and get near the global minimum.
Tuning GBM
- GBMs are highly flexible - many tuning parameters
- It is time consuming to find the optimal combination of hyperparameters
Number of trees
- GBMs often require many trees;
- GBMs can overfit so the goal is to find the optimal number of trees that minimize the loss function of interest with cross validation.
Tuning parameters
Depth of trees
- The number $d$ of splits in each tree, which controls the complexity of the boosted ensemble.
- Often $d=1$ works well, in which case each tree is a stump consisting of a single split. More commonly, $d$ is greater than 1 but it is unlikely that $d>10$ will be required.
Learning rate
- Controls how quickly the algorithm proceeds down the gradient descent.
- Smaller values reduce the chance of overfitting but also increases the time to find the optimal fit.
- This is also called shrinkage.
Tuning parameters (II)
Subsampling
- Controls if a fraction of the available training observations is used.
- Using less than 100% of the training observations means you are implementing stochastic gradient descent.
- This can help to minimize overfitting and keep from getting stuck in a local minimum or plateau of the loss function gradient.
The necessary packages
library(rsample) # data splitting
library(gbm) # basic implementation
library(xgboost) # a faster implementation of gbm
library(caret) # aggregator package - machine learning
library(pdp) # model visualization
library(ggplot2) # model visualization
library(lime) # model visualization
The dataset
- Again, we use the Ames housing dataset
ames_data <- AmesHousing::make_ames()
set.seed(123)
ames_split <- initial_split(ames_data,prop=.7)
ames_train <- training(ames_split)
ames_test <- testing(ames_split)
Package implementation
The most popular implementations of GBM in R:
gbm
The original R implementation of GBMs
xgboost
A fast and efficient gradient boosting framework (C++ backend).
h2o
A powerful java-based interface that provides parallel distributed algorithms and efficient productionalization.
The R-package gbm
- The
gbmR package is an implementation of extensions to Freund and Schapire’s AdaBoost algorithm and Friedman’s gradient boosting machine.
Basic implementation - training function
- Two primary training functions are available:
gbm::gbmandgbm::gbm.fit. gbm::gbmuses the formula interface to specify the modelgbm::gbm.fitrequires the separated x and y matrices (more efficient with many variables).
- The default settings in
gbminclude a learning rate (shrinkage) of 0.001. - This is a very small learning rate and typically requires a large number of trees to find the minimum MSE.
gbmuses the default number of 100 trees, which is rarely sufficient.- The default depth of each tree (
interaction.depth) is 1, which means we are ensembling a bunch of stumps. - We will use
cv.foldsto perform a 5 fold cross validation. - The model takes about 90 seconds to run and the results show that the MSE loss function is minimized with 10000 trees.
Train a GBM model
distribution- depends on the response (e.g. bernoulli for binomial)gaussianis the defaulf value
set.seed(123)
gbm.fit <- gbm(formula = Sale_Price ~ .,distribution="gaussian",
data = ames_train,n.trees = 10000,interaction.depth = 1,
shrinkage = 0.001,cv.folds = 5,n.cores = NULL,verbose = FALSE)
print(gbm.fit) # print results
## gbm(formula = Sale_Price ~ ., distribution = "gaussian", data = ames_train,
## n.trees = 10000, interaction.depth = 1, shrinkage = 0.001,
## cv.folds = 5, verbose = FALSE, n.cores = NULL)
## A gradient boosted model with gaussian loss function.
## 10000 iterations were performed.
## The best cross-validation iteration was 10000.
## There were 80 predictors of which 45 had non-zero influence.
Exercise
- Take some time to dig around in the
gbm.fitobject to get comfortable with its components.
The output object…
- … is a list containing several modelling and results information.
- We can access this information with regular indexing;
- The minimum CV RMSE is 29133 (this means on average our model is about $29,133 off from the actual sales price) but the plot also illustrates that the CV error is still decreasing at 10,000 trees.
Get MSE
sqrt(min(gbm.fit$cv.error))
## [1] 29133.33
Plot loss function as a result of n trees added to the ensemble
gbm.perf(gbm.fit, method = "cv")

## [1] 10000
Tuning GBMs
- The learning rate is increased to take larger steps down the gradient descent,
- The number of trees is reduced (since we reduced the learning rate), and increase the depth of each tree.
set.seed(123)
gbm.fit2 <- gbm(formula = Sale_Price ~ .,
distribution = "gaussian",data = ames_train,
n.trees = 5000,interaction.depth = 3,shrinkage = 0.1,
cv.folds = 5,n.cores = NULL,verbose = FALSE)
# find index for n trees with minimum CV error
min_MSE <- which.min(gbm.fit2$cv.error)
# get MSE and compute RMSE
sqrt(gbm.fit2$cv.error[min_MSE])
## [1] 23112.1
plot loss function as a result of n trees added to the ensemble
Assess the GBM performance:
gbm.perf(gbm.fit2, method = "cv")

## [1] 1260
Grid search
n.minobsinnodeis the minimum number of observations allowed in the trees (nr. for terminal nodes is varied)
hyper_grid <- expand.grid(
shrinkage = c(.01, .1, .3),
interaction.depth = c(1, 3, 5),
n.minobsinnode = c(5, 10, 15),
bag.fraction = c(.65, .8, 1),
optimal_trees = 0,# a place to dump results
min_RMSE = 0
)
# total number of combinations
nrow(hyper_grid)
## [1] 81
Randomize data
train.fractionuse the first XX% of the data so its important to randomize the rows in case there is any logic ordering (i.e. ordered by neighborhood).
random_index <- sample(1:nrow(ames_train), nrow(ames_train))
random_ames_train <- ames_train[random_index, ]
Grid search - loop over hyperparameter grid
for(i in 1:nrow(hyper_grid)) {
set.seed(123)
gbm.tune <- gbm(
formula = Sale_Price ~ .,distribution = "gaussian",
data = random_ames_train,n.trees = 5000,
interaction.depth = hyper_grid$interaction.depth[i],
shrinkage = hyper_grid$shrinkage[i],
n.minobsinnode = hyper_grid$n.minobsinnode[i],
bag.fraction = hyper_grid$bag.fraction[i],
train.fraction = .75,n.cores = NULL,verbose = FALSE
)
# add min training error and trees to grid
hyper_grid$optimal_trees[i] <- which.min(gbm.tune$valid.error)
hyper_grid$min_RMSE[i] <- sqrt(min(gbm.tune$valid.error))
}
The top 10 values
hyper_grid %>%
dplyr::arrange(min_RMSE) %>%
head(10)
Loop through hyperparameter combinations
- We loop through each hyperparameter combination (5,000 trees).
- To speed up the tuning process, instead of performing 5-fold CV we train on 75% of the training observations and evaluate performance on the remaining 25%.
- The top model has better performance than our previously fitted model, with a RMSE nearly $3,000 and lower.
A look at the top 10 models:
- None of the top models used a learning rate of 0.3; small incremental steps down the gradient descent work best,
- None of the top models used stumps (
interaction.depth = 1); there are likely some important interactions that the deeper trees are able to capture. - Adding a stochastic component with
bag.fraction< 1 seems to help; there may be some local minimas in our loss function gradient,
Refine the search - adjust the grid
# modify hyperparameter grid
hyper_grid <- expand.grid(
shrinkage = c(.01, .05, .1),
interaction.depth = c(3, 5, 7),
n.minobsinnode = c(5, 7, 10),
bag.fraction = c(.65, .8, 1),
optimal_trees = 0,# a place to dump results
min_RMSE = 0# a place to dump results
)
# total number of combinations
nrow(hyper_grid)
## [1] 81
The final model
set.seed(123)
# train GBM model
gbm.fit.final <- gbm(formula = Sale_Price ~ .,
distribution = "gaussian",data = ames_train,
n.trees = 483,interaction.depth = 5,
shrinkage = 0.1,n.minobsinnode = 5,
bag.fraction = .65,train.fraction = 1,
n.cores = NULL, # will use all cores by default
verbose = FALSE)
Visualizing - Variable importance
cBarsallows you to adjust the number of variables to show
summary(gbm.fit.final,cBars = 10,
# also can use permutation.test.gbm
method = relative.influence,las = 2)

## var rel.inf
## Overall_Qual Overall_Qual 4.048861e+01
## Gr_Liv_Area Gr_Liv_Area 1.402976e+01
## Neighborhood Neighborhood 1.320178e+01
## Total_Bsmt_SF Total_Bsmt_SF 5.227608e+00
## Garage_Cars Garage_Cars 3.296480e+00
## First_Flr_SF First_Flr_SF 2.927688e+00
## Garage_Area Garage_Area 2.779194e+00
## Exter_Qual Exter_Qual 1.618451e+00
## Second_Flr_SF Second_Flr_SF 1.517907e+00
## Lot_Area Lot_Area 1.296325e+00
## Bsmt_Qual Bsmt_Qual 1.106896e+00
## MS_SubClass MS_SubClass 9.788950e-01
## Bsmt_Unf_SF Bsmt_Unf_SF 8.313946e-01
## Year_Remod_Add Year_Remod_Add 7.381023e-01
## BsmtFin_Type_1 BsmtFin_Type_1 6.967389e-01
## Overall_Cond Overall_Cond 6.939359e-01
## Kitchen_Qual Kitchen_Qual 6.649051e-01
## Garage_Type Garage_Type 5.727906e-01
## Fireplace_Qu Fireplace_Qu 5.281111e-01
## Bsmt_Exposure Bsmt_Exposure 4.601763e-01
## Sale_Type Sale_Type 4.063181e-01
## Exterior_1st Exterior_1st 3.889487e-01
## Open_Porch_SF Open_Porch_SF 3.797533e-01
## Sale_Condition Sale_Condition 3.771138e-01
## Exterior_2nd Exterior_2nd 3.482380e-01
## Year_Built Year_Built 3.289944e-01
## Full_Bath Full_Bath 3.070894e-01
## Screen_Porch Screen_Porch 2.907584e-01
## Fireplaces Fireplaces 2.906831e-01
## Bsmt_Full_Bath Bsmt_Full_Bath 2.890666e-01
## Wood_Deck_SF Wood_Deck_SF 2.448412e-01
## Longitude Longitude 2.249308e-01
## Central_Air Central_Air 2.232338e-01
## Condition_1 Condition_1 1.867403e-01
## Latitude Latitude 1.650208e-01
## Lot_Frontage Lot_Frontage 1.488962e-01
## Heating_QC Heating_QC 1.456177e-01
## Mo_Sold Mo_Sold 1.406729e-01
## TotRms_AbvGrd TotRms_AbvGrd 1.162828e-01
## Functional Functional 1.118959e-01
## Mas_Vnr_Area Mas_Vnr_Area 1.005664e-01
## Paved_Drive Paved_Drive 9.346526e-02
## Land_Contour Land_Contour 9.026977e-02
## Enclosed_Porch Enclosed_Porch 7.512751e-02
## Garage_Cond Garage_Cond 7.143649e-02
## Lot_Config Lot_Config 6.355982e-02
## Year_Sold Year_Sold 6.198938e-02
## Bedroom_AbvGr Bedroom_AbvGr 6.146463e-02
## Mas_Vnr_Type Mas_Vnr_Type 5.820946e-02
## Roof_Style Roof_Style 5.682478e-02
## Roof_Matl Roof_Matl 4.948087e-02
## Alley Alley 4.279173e-02
## BsmtFin_Type_2 BsmtFin_Type_2 3.482743e-02
## Low_Qual_Fin_SF Low_Qual_Fin_SF 3.427532e-02
## Garage_Qual Garage_Qual 3.136598e-02
## Foundation Foundation 2.803887e-02
## Bsmt_Cond Bsmt_Cond 2.768063e-02
## Condition_2 Condition_2 2.751263e-02
## Bsmt_Half_Bath Bsmt_Half_Bath 2.650291e-02
## MS_Zoning MS_Zoning 2.346874e-02
## Pool_QC Pool_QC 2.211997e-02
## Half_Bath Half_Bath 2.148830e-02
## Three_season_porch Three_season_porch 1.833322e-02
## BsmtFin_SF_2 BsmtFin_SF_2 1.774457e-02
## Garage_Finish Garage_Finish 1.407325e-02
## Fence Fence 1.188101e-02
## House_Style House_Style 1.105981e-02
## Exter_Cond Exter_Cond 1.061516e-02
## Misc_Val Misc_Val 7.996187e-03
## Street Street 7.460435e-03
## Land_Slope Land_Slope 6.847183e-03
## Lot_Shape Lot_Shape 6.681489e-03
## Electrical Electrical 5.498046e-03
## BsmtFin_SF_1 BsmtFin_SF_1 4.149129e-03
## Pool_Area Pool_Area 3.622225e-03
## Misc_Feature Misc_Feature 7.232842e-04
## Utilities Utilities 0.000000e+00
## Bldg_Type Bldg_Type 0.000000e+00
## Heating Heating 0.000000e+00
## Kitchen_AbvGr Kitchen_AbvGr 0.000000e+00
Variable importance
Partial dependence plots
- PDPs show the marginal effect one or two features have on the predicted outcome.
- The following PDP plot displays the average change in predicted sales price as we vary
Gr_Liv_Areawhile holding all other variables constant. <!– - This is done by holding all variables constant for each observation in our training data set but then apply the unique values of Gr_Liv_Area for each observation. –>
- We then average the sale price across all the observations.
- This PDP illustrates how the predicted sales price increases as the square footage of the ground floor in a house increases.
Partial dependence plot - Gr_Liv_Area
gbm.fit.final %>% partial(pred.var = "Gr_Liv_Area",
n.trees = gbm.fit.final$n.trees,
grid.resolution = 100) %>%
autoplot(rug = TRUE, train = ames_train) +
scale_y_continuous(labels = scales::dollar)
Partial dependence plot

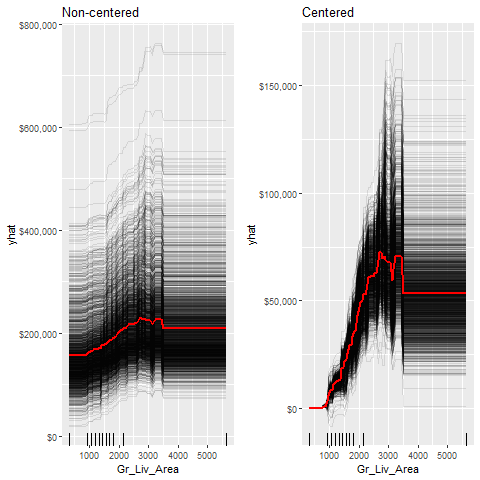
Individual Conditional Expectation (ICE) curves …
- … are an extension of PDP plots but the change in the predicted response variable is plotted as we vary each predictor variable. <!–
- Below shows the regular ICE curve plot (left) and the centered ICE curves (right). –>
- When the curves have a wide range of intercepts and are consequently “stacked” on each other, heterogeneity in the response variable values due to marginal changes in the predictor variable of interest can be difficult to discern.
- The centered ICE can help draw these inferences out and can highlight any strong heterogeneity in our results.
- The results show that most observations follow a common trend as
Gr_Liv_Areaincreases; - the centered ICE plot highlights a few observations that deviate from the common trend.
Non centered ICE curve
ice1 <- gbm.fit.final %>%
partial(
pred.var = "Gr_Liv_Area",
n.trees = gbm.fit.final$n.trees,
grid.resolution = 100,
ice = TRUE
) %>%
autoplot(rug = TRUE, train = ames_train, alpha = .1) +
ggtitle("Non-centered") +
scale_y_continuous(labels = scales::dollar)
Centered ICE curve
ice2 <- gbm.fit.final %>%
partial(
pred.var = "Gr_Liv_Area",
n.trees = gbm.fit.final$n.trees,
grid.resolution = 100,
ice = TRUE
) %>%
autoplot(rug = TRUE, train = ames_train, alpha = .1,
center = TRUE) + ggtitle("Centered") +
scale_y_continuous(labels = scales::dollar)
Non centered and centered ice curve
gridExtra::grid.arrange(ice1, ice2, nrow = 1)
 {height=80%}
{height=80%}
Local Interpretable Model-Agnostic Explanations – (LIME)
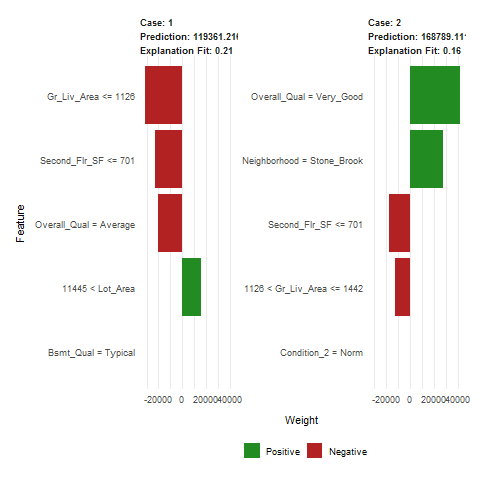
- LIME is a newer procedure for understanding why a prediction resulted in a given value for a single observation.
- To use the
limepackage on agbmmodel we need to define model type and prediction methods.
model_type.gbm <- function(x, ...) {
return("regression")
}
predict_model.gbm <- function(x, newdata, ...) {
pred <- predict(x, newdata, n.trees = x$n.trees)
return(as.data.frame(pred))
}
Applying LIME
- The results show the predicted value (Case 1: 118K Dollar, Case 2: 161K Dollar), local model fit (both are relatively poor), and the most influential variables driving the predicted value for each observation.
# get a few observations to perform local interpretation on
local_obs <- ames_test[1:2, ]
# apply LIME
explainer <- lime(ames_train, gbm.fit.final)
explanation <- explain(local_obs, explainer, n_features = 5)
LIME plot
plot_features(explanation)
Predicting
- If you have decided on a final model you’ll likely want to use the model to predict on new observations.
- Like most models, we simply use the predict function; we also need to supply the number of trees to use (see
?predict.gbmfor details). - The RMSE for the test set is very close to the RMSE we obtained on our best gbm model.
# predict values for test data
pred <- predict(gbm.fit.final, n.trees = gbm.fit.final$n.trees,
ames_test)
# results
caret::RMSE(pred, ames_test$Sale_Price)
## [1] 21533.65
xgboost
- The
xgboostR package provides an R API to “Extreme Gradient Boosting”, which is an efficient implementation of gradient boosting framework (approx. 10x faster than gbm). - The xgboost/demo repository provides a wealth of information. <!– You can also find a fairly comprehensive parameter tuning guide here.
https://www.analyticsvidhya.com/blog/2016/03/complete-guide-parameter-tuning-xgboost-with-codes-python/ –> <!–
- The
xgboostpackage has been quite popular and successful on Kaggle for data mining competitions. –>
Features include:
- Provides built-in k-fold cross-validation -Stochastic GBM with column and row sampling (per split and per tree) for better generalization.
- Includes efficient linear model solver and tree learning algorithms.
- Parallel computation on a single machine.
- Supports various objective functions, including regression, classification and ranking.
- The package is made to be extensible, so that users are also allowed to define their own objectives easily.
- Apache 2.0 License.
Basic implementation
XGBoostonly works with matrices that contain all numeric variables; consequently, we need to hot encode our data. There are different ways to do this in R (i.e.Matrix::sparse.model.matrix,caret::dummyVars) but here we will use thevtreatpackage.vtreatis a robust package for data prep and helps to eliminate problems caused by missing values, novel categorical levels that appear in future data sets that were not in the training data, etc.vtreatis not very intuitive.
Application of vtreat to one-hot encode the training and testing data sets.
# variable names
features <- setdiff(names(ames_train), "Sale_Price")
# Create the treatment plan from the training data
treatplan <- vtreat::designTreatmentsZ(ames_train, features,
verbose = FALSE)
# Get the "clean" variable names from the scoreFrame
new_vars <- treatplan %>%
magrittr::use_series(scoreFrame) %>%
dplyr::filter(code %in% c("clean", "lev")) %>%
magrittr::use_series(varName)
# Prepare the training data
features_train <- vtreat::prepare(treatplan, ames_train,
varRestriction = new_vars) %>% as.matrix()
response_train <- ames_train$Sale_Price
Prepare the test data
features_test <- vtreat::prepare(treatplan, ames_test,
varRestriction = new_vars) %>% as.matrix()
response_test <- ames_test$Sale_Price
dimensions of one-hot encoded data
dim(features_train)
## [1] 2051 343
dim(features_test)
## [1] 879 343
xgboost - training functions
xgboostprovides different training functions (i.e.xgb.trainwhich is just a wrapper forxgboost).-
To train an XGBoost we typically want to use
xgb.cv, which incorporates cross-validation. The following trains a basic 5-fold cross validated XGBoost model with 1,000 trees. There are many parameters available inxgb.cvbut the ones used in this tutorial include the following default values: - learning rate ($\eta$): 0.3
- tree depth (
max_depth): 6 - minimum node size (
min_child_weight): 1 - percent of training data to sample for each tree (subsample –> equivalent to gbm’s
bag.fraction): 100%
Extreme gradient boosting for regression models
set.seed(123)
xgb.fit1 <- xgb.cv(
data = features_train,
label = response_train,
nrounds = 1000, nfold = 5,
objective = "reg:linear", # for regression models
verbose = 0 # silent,
)
get number of trees that minimize error
- The
xgb.fit1object contains lots of good information. - In particular we can assess the
xgb.fit1$evaluation_logto identify the minimum RMSE and the optimal number of trees for both the training data and the cross-validated error. - The training error continues to decrease to 924 trees where the RMSE nearly reaches zero;
- The cross validated error reaches a minimum RMSE of 27,337 with only 60 trees.
xgb.fit1$evaluation_log %>%
dplyr::summarise(
ntrees.train=which(train_rmse_mean==min(train_rmse_mean))[1],
rmse.train= min(train_rmse_mean),
ntrees.test=which(test_rmse_mean==min(test_rmse_mean))[1],
rmse.test = min(test_rmse_mean)
)
## ntrees.train rmse.train ntrees.test rmse.test
## 1 924 0.0483002 60 27337.79
Plot error vs number trees
ggplot(xgb.fit1$evaluation_log) +
geom_line(aes(iter, train_rmse_mean), color = "red") +
geom_line(aes(iter, test_rmse_mean), color = "blue")

Early stopping
- A nice feature provided by
xgb.cvis early stopping. - This allows us to tell the function to stop running if the cross validated error does not improve for n continuous trees.
- E.g., the above model could be re-run with the following where we tell it stop if we see no improvement for 10 consecutive trees. This feature will help us speed up the tuning process.
set.seed(123)
xgb.fit2 <- xgb.cv(data = features_train,
label = response_train,
nrounds = 1000, nfold = 5,
objective = "reg:linear", # for regression models
verbose = 0, # silent,
# stop if no improvement for 10 consecutive trees
early_stopping_rounds = 10)
plot error vs number trees
ggplot(xgb.fit2$evaluation_log) +
geom_line(aes(iter, train_rmse_mean), color = "red") +
geom_line(aes(iter, test_rmse_mean), color = "blue")

Tuning
-
To tune the XGBoost model we pass parameters as a list object to the params argument. The most common parameters include:
eta:controls- the learning ratemax_depth: tree depthmin_child_weight: minimum number of observations required in each terminal nodesubsample: percent of training data to sample for each treecolsample_bytrees: percent of columns to sample from for each tree- E.g. to specify specific values for these parameters we would extend the above model with the following parameters.
create parameter list
params <- list(
eta = .1,
max_depth = 5,
min_child_weight = 2,
subsample = .8,
colsample_bytree = .9
)
To perform a large search grid,…
- we can follow the same procedure we did with
gbm. - We create our hyperparameter search grid along with columns to dump our results in.
- Here, we have a pretty large search grid consisting of 576 different hyperparameter combinations to model.
# create hyperparameter grid
hyper_grid <- expand.grid(
eta = c(.01, .05, .1, .3),
max_depth = c(1, 3, 5, 7),
min_child_weight = c(1, 3, 5, 7),
subsample = c(.65, .8, 1),
colsample_bytree = c(.8, .9, 1),
optimal_trees = 0,# a place to dump results
min_RMSE = 0# a place to dump results
)
nrow(hyper_grid)
## [1] 576
train model
set.seed(123)
xgb.fit3 <- xgb.cv(
params = params,
data = features_train,
label = response_train,
nrounds = 1000,
nfold = 5,
objective = "reg:linear", # for regression models
verbose = 0, # silent,
# stop if no improvement for 10 consecutive trees
early_stopping_rounds = 10
)
assess results
xgb.fit3$evaluation_log %>%
dplyr::summarise(
ntrees.train=which(train_rmse_mean==min(train_rmse_mean))[1],
rmse.train= min(train_rmse_mean),
ntrees.test= which(test_rmse_mean==min(test_rmse_mean))[1],
rmse.test= min(test_rmse_mean)
)
## ntrees.train rmse.train ntrees.test rmse.test
## 1 211 5222.229 201 24411.64
Loop through a XGBoost model
- We apply the same in the loop and apply a
XGBoostmodel for each hyperparameter combination and dump the results in thehyper_griddata frame.
Important note:
- If you plan to run this code be prepared to run it before going out to eat or going to bed as it the full search grid took 6 hours to run!
Grid search
for(i in 1:nrow(hyper_grid)) {
params <- list(# create parameter list
eta = hyper_grid$eta[i],max_depth = hyper_grid$max_depth[i],
min_child_weight = hyper_grid$min_child_weight[i],
subsample = hyper_grid$subsample[i],
colsample_bytree = hyper_grid$colsample_bytree[i])
set.seed(123)
xgb.tune <- xgb.cv(params = params,data = features_train,
label = response_train,nrounds=5000,nfold=5,objective = "reg:linear",
#stop if no improvement for 10 consecutive trees
verbose = 0,early_stopping_rounds = 10 )
# add min training error and trees to grid
hyper_grid$optimal_trees[i]<-which.min(
xgb.tune$evaluation_log$test_rmse_mean)
hyper_grid$min_RMSE[i] <- min(
xgb.tune$evaluation_log$test_rmse_mean)
}
Result - top 10 models
hyper_grid %>%
dplyr::arrange(min_RMSE) %>%
head(10)
## eta max_depth min_child_weight subsample colsample_bytree
## 1 0.01 1 1 0.65 0.8
## 2 0.05 1 1 0.65 0.8
## 3 0.10 1 1 0.65 0.8
## 4 0.30 1 1 0.65 0.8
## 5 0.01 3 1 0.65 0.8
## 6 0.05 3 1 0.65 0.8
## 7 0.10 3 1 0.65 0.8
## 8 0.30 3 1 0.65 0.8
## 9 0.01 5 1 0.65 0.8
## 10 0.05 5 1 0.65 0.8
## optimal_trees min_RMSE
## 1 0 0
## 2 0 0
## 3 0 0
## 4 0 0
## 5 0 0
## 6 0 0
## 7 0 0
## 8 0 0
## 9 0 0
## 10 0 0
The top model
- After assessing the results you would likely perform a few more grid searches to hone in on the parameters that appear to influence the model the most. <!–
- Here is a link to a great blog post that discusses a strategic approach to tuning with
xgboost. –> - We’ll just assume the top model in the above search is the globally optimal model. Once you’ve found the optimal model, we can fit our final model with
xgb.train.
# parameter list
params <- list(
eta = 0.01,
max_depth = 5,
min_child_weight = 5,
subsample = 0.65,
colsample_bytree = 1
)
Train final model
xgb.fit.final <- xgboost(
params = params,
data = features_train,
label = response_train,
nrounds = 1576,
objective = "reg:linear",
verbose = 0
)
Input types for xgboost
Input type: xgboost takes several types of input data:
- Dense Matrix: R’s dense matrix, i.e. matrix ;
- Sparse Matrix: R’s sparse matrix, i.e. Matrix::dgCMatrix ;
- Data File: local data files ;
xgb.DMatrix: its own class (recommended).
get information
- We get information on an
xgb.DMatrixobject withgetinfo
Visualizing
Variable importance
xgboost provides built-in variable importance plotting. First, you need to create the importance matrix with xgb.importance and then feed this matrix into xgb.plot.importance. There are 3 variable importance measure:
- Gain: the relative contribution of the corresponding feature to the model calculated by taking each feature’s contribution for each tree in the model. This is synonymous with gbm’s relative.influence.
- Cover: the relative number of observations related to this feature. For example, if you have 100 observations, 4 features and 3 trees, and suppose feature1 is used to decide the leaf node for 10, 5, and 2 observations in tree1, tree2 and tree3 respectively; then the metric will count cover for this feature as 10+5+2 = 17 observations. This will be calculated for all the 4 features and the cover will be 17 expressed as a percentage for all features’ cover metrics.
create importance matrix
- Frequency: the percentage representing the relative number of times a particular feature occurs in the trees of the model. In the above example, if feature1 occurred in 2 splits, 1 split and 3 splits in each of tree1, tree2 and tree3; then the weightage for feature1 will be 2+1+3 = 6. The frequency for feature1 is calculated as its percentage weight over weights of all features.
importance_matrix <- xgb.importance(model = xgb.fit.final)
variable importance plot
xgb.plot.importance(importance_matrix, top_n = 10, measure = "Gain")
Variable importance plot

LIME
- LIME provides built-in functionality for xgboost objects (see ?model_type).
- Just keep in mind that the local observations being analyzed need to be one-hot encoded in the same manner as we prepared the training and test data. Also, when you feed the training data into the lime::lime function be sure that you coerce it from a matrix to a data frame.
# one-hot encode the local observations to be assessed.
local_obs_onehot <- vtreat::prepare(treatplan, local_obs,
varRestriction = new_vars)
# apply LIME
explainer <- lime(data.frame(features_train), xgb.fit.final)
explanation <- explain(local_obs_onehot, explainer,
n_features = 5)
Plot the features
plot_features(explanation)

Predicting on new observations
unlike GBM we do not need to provide the number of trees. Our test set RMSE is only about $600 different than that produced by our gbm model.
# predict values for test data
pred <- predict(xgb.fit.final, features_test)
# results
caret::RMSE(pred, response_test)
## [1] 21898
## [1] 21319.3