Random Forests
- Bagging can turn a single tree model with high variance and poor predictive power into a fairly accurate prediction function.
- But bagging suffers from tree correlation, which reduces the overall performance of the model.
- Random forests are a modification of bagging that builds a large collection of de-correlated trees
- It is a very popular out-of-the-box learning algorithm that enjoys good predictive performance.
Extending the bagging technique
- Bagging introduces a random component in to the tree building process
- The trees in bagging are not completely independent of each other since all the original predictors are considered at every split of every tree.
- Trees from different bootstrap samples have similar structure to each other (especially at the top of the tree) due to underlying relationships.
Similar trees - tree correlation
- If we create six decision trees with different bootstrapped samples of the Boston housing data, the top of the trees all have a very similar structure.
- Although there are 15 predictor variables to split on, all six trees have both
lstatandrmvariables driving the first few splits.
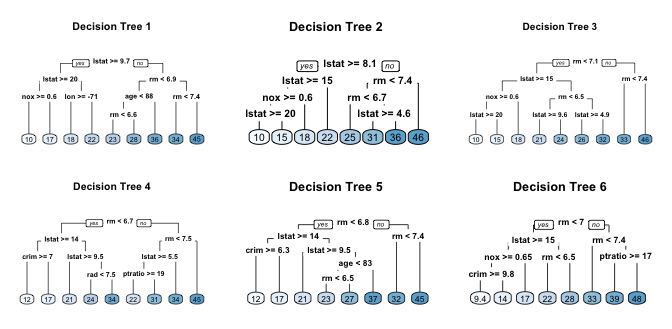
Tree correlation
- Tree correlation prevents bagging from optimally reducing variance of the predictive values.
- To reduce variance further, we need to minimize the amount of correlation between the trees.
- This can be achieved by injecting more randomness into the tree-growing process.
Random forests achieve this in two ways:
1) Bootstrap:
- Similar to bagging, each tree is grown to a bootstrap resampled data set, which makes them different and decorrelates them.
2) Split-variable randomization:
- For every split, the search for the split variable is limited to a random subset of $m$ of the $p$ variables.
- For regression trees, typical default values are $m=p/3$ (tuning parameter).
- When $m=p$, the randomization is limited (only step 1) and is the same as bagging.
Basic algorithm
The basic algorithm for a regression random forest can be generalized:
1. Given training data set
2. Select number of trees to build (ntrees)
3. for i = 1 to ntrees do
4. | Generate a bootstrap sample of the original data
5. | Grow a regression tree to the bootstrapped data
6. | for each split do
7. | | Select m variables at random from all p variables
8. | | Pick the best variable/split-point among the m
9. | | Split the node into two child nodes
10. | end
11. | Use tree model stopping criteria to determine: tree complete
12. end
The algorithm randomly selects a bootstrap sample to train and predictors to use at each split.
Characteristics
- Since bootstrap samples and features are selected randomly at each split, we create a more diverse set of trees, which tends to lessen tree correlation beyond bagged trees and often dramatically increase predictive power. <!–
- random forests have the least variability in their prediction accuracy when tuning. –>
out-of-bag error
- Similar to bagging, a natural benefit of the bootstrap resampling process is that random forests have an out-of-bag (OOB) sample that provides an efficient and reasonable approximation of the test error.
- This provides a built-in validation set without any extra work, and you do not need to sacrifice any of your training data to use for validation.
- We are more efficient identifying the number of trees required to stablize the error rate <!– during tuning;
Out-of-bag error vs. validation error
- Some difference between the OOB error and test error are expected.
–>
Preparation - random forests
- The following slides are based on UC Business Analytics R Programming Guide on random forests
library(rsample) # data splitting
library(randomForest) # basic implementation
library(ranger) # a faster implementation of randomForest
# an aggregator package for performing many
# machine learning models
library(caret)
The Ames housing data
set.seed(123)
ames_data <- AmesHousing::ames_raw
set.seed(123)
ames_split <- rsample::initial_split(ames_data,prop=.7)
ames_train <- rsample::training(ames_split)
ames_test <- rsample::testing(ames_split)
Basic implementation
- There are over 20 random forest packages in R.
- To demonstrate the basic implementation we use the
randomForestpackage, the oldest and most well known implementation of the random forest algorithm in R. - As your data set grows in size
randomForestdoes not scale well (although you can parallelize withforeach). - To explore and compare a variety of tuning parameters we can find more effective packages.
- The package
rangerwill be presented in the tuning section.
randomForest::randomForest
randomForestcan use the formula or x-y matrix notation.- Below we apply the default
randomForestmodel using the formal specification. - The default random forest performs 500 trees and $\dfrac{\text{nr. features}}{3}=26$ randomly selected predictor variables at each split.
set.seed(123)
# default RF model
(m1 <- randomForest(formula = Sale_Price ~ .,data=ames_train))
##
## Call:
## randomForest(formula = Sale_Price ~ ., data = ames_train)
## Type of random forest: regression
## Number of trees: 500
## No. of variables tried at each split: 26
##
## Mean of squared residuals: 639516350
## % Var explained: 89.7
Plotting the model
- The error rate stabalizes with around 100 trees but continues to decrease slowly until around 300 trees.
plot(m1,main="Error rate")
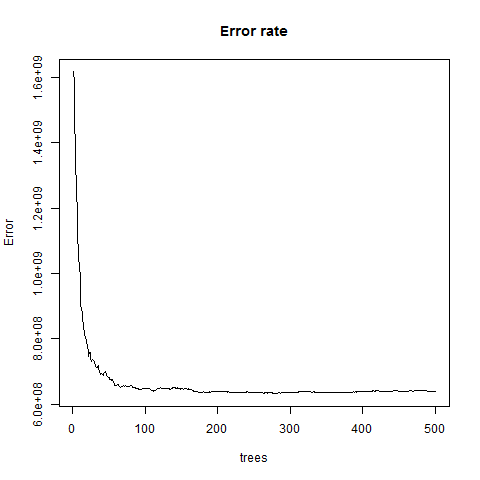 {height=65%}
{height=65%}
Random forests - out-of-the-box algorithm
- Random forests perform remarkably well with very little tuning.
- We get an RMSE of less than 30K dollar without any tuning.
- This is more than 6K dollar RMSE-reduction compared to a fully-tuned bagging model
- and 4K dollar reduction to to a fully-tuned elastic net model.
- We can still seek improvement by tuning our random forest model.
Tuning Random forests
- Random forests are fairly easy to tune since there are only a handful of tuning parameters.
- First we tune the number of candidate variables to select from at each split.
- A few additional hyperparameters are important.
Tuning parameters (I)
- The following hyperparameter are important (names may differ across packages):
number of trees
ntree- We want enough trees to stabalize the error but using too many trees is inefficient, esp. for large data sets.
number of variables
mtry- number of variables as candidates at each split. Whenmtry=pthe model equates to bagging.- When
mtry=1the split variable is completely random, all variables get a chance but can lead to biased results. Suggestion: start with 5 values evenly spaced across the range from 2 to p.
Tuning parameters (II)
Number of samples
sampsize- Default value is 63.25% since this is the expected value of unique observations in the bootstrap sample.- Lower sample sizes can reduce training time but may introduce more bias. Increasing sample size can increase performance but at risk of overfitting - it introduces more variance. <!–
- When tuning this parameter we stay near the 60-80% range. –>
Tuning parameters (III)
minimum number of samples within the terminal nodes:
nodesize- Controls the complexity of the trees.- It is the minimum size of terminal nodes.
- Smaller node size allow for deeper, more complex trees
- This is another bias-variance tradeoff where deeper trees introduce more variance (risk of overfitting)
- Shallower trees introduce more bias (risk of not fully capturing unique patters and relatonships in the data).
maximum number of terminal nodes
maxnodes: A way to control the complexity of the trees.- More nodes equates to deeper, more complex trees.
- Less nodes result in shallower trees.
Initial tuning with randomForest
- If we just tune the
mtryparameter we can userandomForest::tuneRFfor a quick and easy tuning assessment. <!– tuneRfwill start at a value ofmtrythat you supply and increase by a certain step factor until the OOB error stops improving. –>- We start with 5 candidate variables (
mtryStart=5) and increase by a factor of 2 until the OOB error stops improving by 1 per cent. tuneRFrequires a separate x y specification.- The optimal
mtryvalue in this sequence is very close to the default mtry value of $\dfrac{\text{features}}{3}=26$.
features <- setdiff(names(ames_train), "Sale_Price")
set.seed(123)
m2<-tuneRF(x= ames_train[,features],
y= ames_train$Sale_Price,ntreeTry = 500,
mtryStart = 5,stepFactor = 2,
improve = 0.01,trace=FALSE)
Full grid search with ranger
- To perform a larger grid search across several hyperparameters we’ll need to create a grid, loop through each hyperparameter combination and evaluate the model.
- Unfortunately, this is where
randomForestbecomes quite inefficient since it does not scale well. - Instead, we can use
rangerwhich is a C++ implementation of Breiman’s random forest algorithm and is over 6 times faster thanrandomForest.
Assessing the speed
randomForest speed
system.time(
ames_randomForest <- randomForest(
formula = Sale_Price ~ .,
data = ames_train,
ntree = 500,
mtry = floor(length(features) / 3)
)
)
# User System elapsed
# 145.47 0.09 152.48
ranger speed
system.time(
ames_ranger <- ranger(formula=Sale_Price ~ .,
data = ames_train,num.trees = 500,
mtry = floor(length(features) / 3))
)
## user system elapsed
## 9.13 0.03 3.23
The grid search
- To perform the grid search, we construct our grid of hyperparameters.
# hyperparameter grid search
hyper_grid <- expand.grid(
mtry = seq(20, 30, by = 2),
node_size = seq(3, 9, by = 2),
sampe_size = c(.55, .632, .70, .80),
OOB_RMSE = 0
)
- We search across 96 different models with varying
mtry, minimum node size, and sample size.
nrow(hyper_grid) # total number of combinations
## [1] 96
Loop - hyperparameter combination (I)
- We apply 500 trees since our previous example illustrated that 500 was plenty to achieve a stable error rate.
- We set the random number generator seed. This allows us to consistently sample the same observations for each sample size and make the impact of each change clearer.
for(i in 1:nrow(hyper_grid)) {
model <- ranger(formula= Sale_Price ~ .,data= ames_train,
num.trees = 500,mtry= hyper_grid$mtry[i],
min.node.size = hyper_grid$node_size[i],
sample.fraction = hyper_grid$sampe_size[i],
seed = 123)
# add OOB error to grid
hyper_grid$OOB_RMSE[i] <- sqrt(model$prediction.error)
}
The results - samll difference between RMSE
hyper_grid %>% dplyr::arrange(OOB_RMSE) %>% head(10)
## mtry node_size sampe_size OOB_RMSE
## 1 26 3 0.8 25404.60
## 2 28 3 0.8 25405.92
## 3 28 5 0.8 25459.46
## 4 26 5 0.8 25493.80
## 5 30 3 0.8 25528.26
## 6 22 3 0.7 25552.73
## 7 26 9 0.8 25554.31
## 8 28 7 0.8 25578.45
## 9 20 3 0.8 25581.23
## 10 24 3 0.8 25590.73
- Models with slighly larger sample sizes (70-80 per cent) and deeper trees (3-5 observations in terminal node) perform best.
- We get various
mtryvalues in top 10 - not over influential.
Hyperparameter grid search - categorical variables
- We use one-hot encoding for our categorical variables which produces 353 predictor variables versus the 80 we were using above.
# one-hot encode our categorical variables
(one_hot <- dummyVars(~ ., ames_train, fullRank = FALSE))
## Dummy Variable Object
##
## Formula: ~.
## 81 variables, 46 factors
## Variables and levels will be separated by '.'
## A less than full rank encoding is used
Make a dataframe of dummy variable object
ames_train_hot<-predict(one_hot,ames_train)%>%as.data.frame()
Hot encoding and hypergrid
# make ranger compatible names
names(ames_train_hot) <- make.names(names(ames_train_hot),
allow_ = FALSE)
# --> same as above but with increased mtry values
hyper_grid_2 <- expand.grid(
mtry = seq(50, 200, by = 25),
node_size = seq(3, 9, by = 2),
sampe_size = c(.55, .632, .70, .80),
OOB_RMSE = 0
)
The best model
The best random forest model:
- uses columnar categorical variables
mtry= 24,- terminal node size of 5 observations
- sample size of 80%.
How to proceed
- Repeat the model to get a better expectation of error rate.
Random forests with ranger
- The
impuritymeasure is the variance of the responses for regression impurityis a measure for heterogeneity - it measures how well the classes are
<!– https://stats.stackexchange.com/questions/220321/what-is-node-impurity-purity-in-decision-trees-in-plain-english-why-do-we-need
https://people.cs.pitt.edu/~milos/courses/cs2750-Spring03/lectures/class19.pdf –>
OOB_RMSE <- vector(mode = "numeric", length = 100)
for(i in seq_along(OOB_RMSE)) {
optimal_ranger <- ranger(formula= Sale_Price ~ .,
data = ames_train,
num.trees = 500,
mtry = 24,
min.node.size = 5,
sample.fraction = .8,
importance = 'impurity')
OOB_RMSE[i] <- sqrt(optimal_ranger$prediction.error)
}
Variable importance / node impurity
- Node impurity represents how well the trees split the data. There are several impurity measures;
-
Gini index, Entropy and misclassification error are options to measure the node impurity
- We set
importance = 'impurity', which allows us to assess variable importance. - Variable importance is measured by recording the decrease in MSE each time a variable is used as a node split in a tree.
- The remaining error left in predictive accuracy after a node split is known as node impurity. <!– https://medium.com/the-artificial-impostor/feature-importance-measures-for-tree-models-part-i-47f187c1a2c3 http://www.cse.msu.edu/~cse802/DecisionTrees.pdf https://www.cs.indiana.edu/~predrag/classes/2017fallb365/ch4.pdf
http://mason.gmu.edu/~jgentle/csi772/16s/L10_CART_16s.pdf
https://stats.stackexchange.com/questions/158583/what-does-node-size-refer-to-in-the-random-forest –>
- A variable that reduces this impurity is considered more imporant than those variables that do not.
- We accumulate the reduction in MSE for each variable across all the trees and the variable with the greatest accumulated impact is considered the more important.
Plot the variable importance
varimp_ranger <- optimal_ranger$variable.importance
lattice::barchart(sort(varimp_ranger)[1:25],col="royalblue")
- We see that Utilities has the greatest impact in reducing MSE across our trees, followed by
names(sort(varimp_ranger))[2], Low_Qual_Fin_SF, etc.
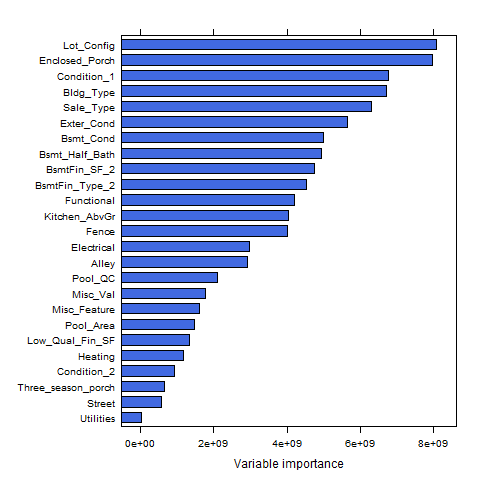 {height=60%}
{height=60%}
A histogram of OOB RMSE
hist(OOB_RMSE, breaks = 20,col="royalblue")
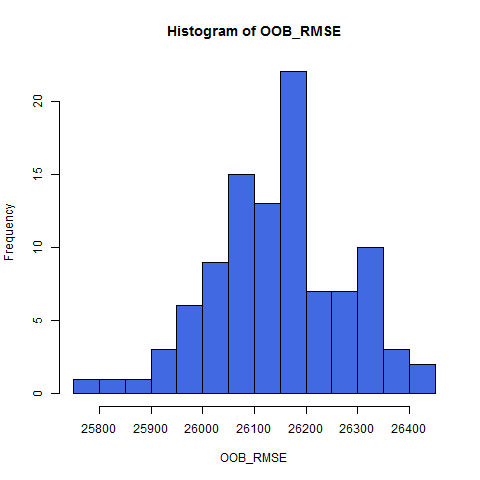 {height=75%}
{height=75%}
Predicting
- With the preferred model we can use the traditional predict function to make predictions on a new data set.
- We can use this for all our model types (
randomForestandranger); although the outputs differ slightly. <!– - not that the new data for the
h2omodel needs to be anh2oobject. –>
# randomForest
pred_randomForest <- predict(ames_randomForest, ames_test)
head(pred_randomForest)
## 1 2 3 4 5 6
## 113543.1 185556.4 259258.1 190943.9 179071.0 480952.3
# ranger
pred_ranger <- predict(ames_ranger, ames_test)
head(pred_ranger$predictions)
## [1] 129258.1 186520.7 265628.2 197745.5 175517.6 392691.7
Summary - random forests
- Random forests provide a very powerful out-of-the-box algorithm that often has great predictive accuracy.
- Because of their more simplistic tuning nature and the fact that they require very little, if any, feature pre-processing they are often one of the first go-to algorithms when facing a predictive modeling problem.
Advantages & Disadvantages
Advantages - random forrests
- Typically have very good performance
- Remarkably good “out-of-the box” - very little tuning required
- Built-in validation set - don’t need to sacrifice data for extra validation
- No pre-processing required
- Robust to outliers
Disadvantages - random forrests
- Can become slow on large data sets
- Although accurate, often cannot compete with advanced boosting algorithms
- Less interpretable
Links
These slides are mainly based on
-
A UC Business Analytics R Programming Guide - section random forests
-
and on the chapter on random forests in the e-book of Brad Boehmke and Brandon Greenwell - Hands-on Machine Learning with R