Insufficient Solution
- When the number of features exceed the number of observations ($p>n$), the OLS solution matrix is not invertible.
- This causes significant issues because it means:
(1) The least-squares estimates are not unique. There are an infinite set of solutions available and most of these solutions overfit the data.
(2) In many instances the result will be computationally infeasible.
- To resolve this issue we can remove variables until $p<n$ and then fit an OLS regression model.
- Although we can use pre-processing tools to apply this manual approach (Kuhn and Johnson, 2013, pp. 43-47), it can be cumbersome and prone to errors.
Regularized Regression
-
When we experience these concerns, one alternative is to use regularized regression (also commonly referred to as penalized models or shrinkage methods) to control the parameter estimates.
-
Regularized regression puts contraints on the magnitude of the coefficients and will progressively shrink them towards zero. This constraint helps to reduce the magnitude and fluctuations of the coefficients and will reduce the variance of our model.
Regularization
elitedatascience.com definition
Regularization is a technique used to prevent overfitting by artificially penalizing model coefficients.
- It can discourage large coefficients (by dampening them).
- It can also remove features entirely (by setting their coefficients to 0).
- The “strength” of the penalty is tunable.
Wikipedia definition of Regularization
Regularization is the process of adding information in order to solve an ill-posed problem or to prevent overfitting.
Strenghts and weaknesses of regularization
Strengths:
- Linear regression is straightforward to understand and explain, and can be regularized to avoid overfitting.
- Linear models can be updated easily with new data
Weaknesses:
- Linear regression in general performs poorly when there are non-linear relationships. - They are not naturally flexible enough to capture more complex patterns, and adding the right interaction terms or polynomials can be tricky and time-consuming.
Three regularized regression algorithms
{ height=40% }
Lasso regression
- Absolute size of coefficients is penalized.
- Coefficients can be exactly 0.
Ridge regression
- Squared size of coefficients is penalized.
- Smaller coefficients, but it doesn’t force them to 0.
Elastic-net
- A mix of both absolute and squared size is penalzied. <!–
- The ratio of the two penalty types should be tuned. –>
The objective function of regularized regression methods…
- is very similar to OLS regression;
- And a penalty parameter (P) is added.
- There are two main penalty parameters, which have a similar effect.
- They constrain the size of the coefficients such that the only way the coefficients can increase is if we experience a comparable decrease in the sum of squared errors (SSE).
Preparations
- Most of the following slides are based on the UC Business Analytics R Programming Guide
Necessary packages
library(rsample) # data splitting
library(glmnet) # implementing regularized regression approaches
library(dplyr) # basic data manipulation procedures
library(ggplot2) # plotting
library(knitr) # for tables
The example dataset
library(AmesHousing)
ames_data <- AmesHousing::make_ames()
Create training (70%) and test (30%) sets
set.seedis used for reproducibilityinitial_splitis used to split data in training and test data
set.seed(123)
ames_split <- rsample::initial_split(ames_data, prop = .7,
strata = "Sale_Price")
ames_train <- rsample::training(ames_split)
ames_test <- rsample::testing(ames_split)
Ridge Regression
-
Ridge regression (Hoerl, 1970) controls the coefficients by adding $\lambda\sum_{j=1}^p\beta_j^2$ to the objective function.
-
This penalty parameter is referred to as “$L_2$” as it signifies a second-order penalty being used on the coefficients.
-
This penalty parameter can take on a wide range of values, which is controlled by the tuning parameter $\lambda$.
-
When $\lambda=0$, there is no effect and our objective function equals the normal OLS regression objective function of simply minimizing SSE.
-
As $\lambda \rightarrow \infty$, the penalty becomes large and forces our coefficients to zero.
Exemplar coefficients
Exemplar coefficients have been regularized with $\lambda$ ranging from 0 to over 8,000 (log(8103)=9).
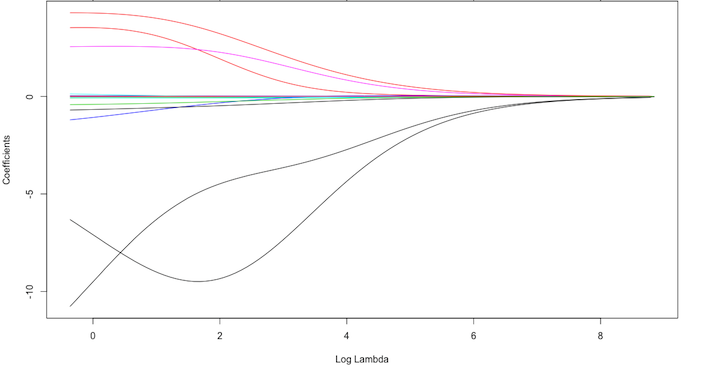
How to choose the right $\lambda$
-
Although these coefficients were scaled and centered prior to the analysis, you will notice that some are extremely large when $\lambda\rightarrow 0$.
-
We have a large negative parameter that fluctuates until $log(\lambda)\approx 2$ where it then continuously shrinks to zero.
-
This is indicates multicollinearity and likely illustrates that constraining our coefficients with $log(\lambda)>2$ may reduce the variance, and therefore the error, in our model.
-
But how do we find the amount of shrinkage (or $\lambda$) that minimizes our error?
Implementation in glmnet
plot(density(ames_data$Sale_Price),main="")

glmnetdoes not use the formula method (y ~ x) so prior to modeling we need to create our feature and target set.- The
model.matrixfunction is used on our feature set, which will automatically dummy encode qualitative variables - We also log transform our response variable due to its skeweness.
Training and testing feature model matrices and response vectors.
- We use
model.matrix(...)[, -1]to discard the intercept
ames_train_x <- model.matrix(Sale_Price ~ ., ames_train)[, -1]
ames_train_y <- log(ames_train$Sale_Price)
ames_test_x <- model.matrix(Sale_Price ~ ., ames_test)[, -1]
ames_test_y <- log(ames_test$Sale_Price)
# What is the dimension of of your feature matrix?
dim(ames_train_x)
## [1] 2054 307
Behind the scenes
- The alpha parameter tells
glmnetto perform a Ridge ($\alpha = 0$), Lasso ($\alpha = 1$), or Elastic Net $(0\leq \alpha \leq 1)$ model. - Behind the scenes,
glmnetis doing two things that you should be aware of:
(1.) It is essential that predictor variables are standardized when performing regularized regression. glmnet performs this for you. If you standardize your predictors prior to glmnet you can turn this argument off with standardize=FALSE.
(2.) glmnet will perform Ridge models across a wide range of $\lambda$ parameters, which are illustrated in the figure on the next slide.
ames_ridge <- glmnet(x = ames_train_x,y = ames_train_y,
alpha = 0)
A wide range of $\lambda$ parameters
plot(ames_ridge, xvar = "lambda")

$\lambda$ values in glmnet
- We can see the exact $\lambda$ values applied with
ames_ridge$lambda. - You can specify your own $\lambda$ values,
- By default
glmnetapplies 100 $\lambda$ values that are data derived. - Normally you will have little need to adjust the default $\lambda$ values.
head(ames_ridge$lambda)
## [1] 289.0010 263.3270 239.9337 218.6187 199.1972 181.5011
Access the coefficients with coef.
- The coefficients are stored for each model in order of largest to smallest $\lambda$.
- The coefficients for the
Gr_Liv_AreaandTotRms_AbvGrdfeatures for the largest $\lambda$ (279.1035) and smallest $\lambda$ (0.02791035) are visible. - The largest $\lambda$ value has pushed these coefficients to nearly 0.
coef(ames_ridge)[c("Gr_Liv_Area", "TotRms_AbvGrd"),100]
## Gr_Liv_Area TotRms_AbvGrd
## 0.0001108687 0.0083032186
coef(ames_ridge)[c("Gr_Liv_Area", "TotRms_AbvGrd"), 1]
## Gr_Liv_Area TotRms_AbvGrd
## 5.848028e-40 1.341550e-37
- But how much improvement we are experiencing in our model.
Tuning
- Recall that $\lambda$ is a tuning parameter that helps to control our model from over-fitting to the training data.
- To identify the optimal $\lambda$ value we need to perform cross-validation (CV).
cv.glmnetprovides a built-in option to perform k-fold CV, and by default, performs 10-fold CV.
ames_ridge <- cv.glmnet(x = ames_train_x,y = ames_train_y,
alpha = 0)
Results of cv Ridge regression
plot(ames_ridge)

- The plot illustrates the 10-fold CV mean squared error (MSE) across the $\lambda$ values.
- We see no substantial improvement;
The plot explained (I)
- As we constrain our coefficients with $log(\lambda)\leq 0$ penalty, the MSE rises considerably.
- The numbers at the top of the plot (301) just refer to the number of variables in the model.
- Ridge regression does not force any variables to exactly zero so all features will remain in the model.
The plot explained (II)
min(ames_ridge$cvm) # minimum MSE
## [1] 0.01955871
ames_ridge$lambda.min # lambda for this min MSE
## [1] 0.1542312
# 1 st.error of min MSE
ames_ridge$cvm[ames_ridge$lambda == ames_ridge$lambda.1se]
## [1] 0.02160821
ames_ridge$lambda.1se # lambda for this MSE
## [1] 0.5169216
The plot explained (III)
- The advantage of identifying the $\lambda$ with an MSE within one standard error becomes more obvious with the Lasso and Elastic Net models.
- For now we can assess this visually.
- We plot the coefficients across the $\lambda$ values and the dashed red line represents the largest $\lambda$ that falls within one standard error of the minimum MSE.
- This shows you how much we can constrain the coefficients while still maximizing predictive accuracy.
ames_ridge_min <- glmnet(x = ames_train_x,y = ames_train_y,
alpha = 0)
Coefficients across the $\lambda$ values
plot(ames_ridge_min, xvar = "lambda")
abline(v = log(ames_ridge$lambda.1se), col = "red",
lty = "dashed")

Advantages and Disadvantages
- The Ridge regression model has pushed many of the correlated features towards each other rather than allowing for one to be wildly positive and the other wildly negative.
- Many of the non-important features have been pushed closer to zero.
- We have reduced the noise in our data $\Rightarrow$ more clarity in identifying the true signals.
coef(ames_ridge, s = "lambda.1se") %>%
filter(row != "(Intercept)") %>%
top_n(25, wt = abs(value)) %>%
ggplot(aes(value, reorder(row, value))) +
geom_point() +
ggtitle("Top 25 influential variables") +
xlab("Coefficient") +
ylab(NULL)
Top 25 influential variables
Exercise: ridge regression (I)
1) Load the lars package and the diabetes dataset
2) Load the glmnet package to implement ridge regression.
The dataset has three matrices x, x2 and y. x has a smaller set of independent variables while x2 contains the full set with quadratic and interaction terms. y is the dependent variable which is a quantitative measure of the progression of diabetes.
3) Generate separate scatterplots with the line of best fit for all the predictors in x with y on the vertical axis.
4) Regress y on the predictors in x using OLS. We will use this result as benchmark for comparison.
Exercise: ridge regression (II)
5) Fit the ridge regression model using the glmnet function and plot the trace of the estimated coefficients against lambdas. Note that coefficients are shrunk closer to zero for higher values of lambda.
6) Use the cv.glmnet function to get the cross validation curve and the value of lambda that minimizes the mean cross validation error.
7) Using the minimum value of lambda from the previous exercise, get the estimated beta matrix. Note that coefficients are lower than least squares estimates.
8) To get a more parsimonious model we can use a higher value of lambda that is within one standard error of the minimum. Use this value of lambda to get the beta coefficients. Note the shrinkage effect on the estimates.
Exercise: ridge regression (III)
9) Split the data randomly between a training set (80%) and test set (20%). We will use these to get the prediction standard error for least squares and ridge regression models.
10) Fit the ridge regression model on the training and get the estimated beta coefficients for both the minimum lambda and the higher lambda within 1-standard error of the minimum.
11) Get predictions from the ridge regression model for the test set and calculate the prediction standard error. Do this for both the minimum lambda and the higher lambda within 1-standard error of the minimum.
12) Fit the least squares model on the training set.
13) Get predictions from the least squares model for the test set and calculate the prediction standard error.
Ridge and Lasso
A Ridge model…
- … is good if we need to retain all features, yet reduce the noise that less influential variables may create and minimize multicollinearity.
- … does not perform feature selection. If greater interpretation is necessary where you need to reduce the signal in your data to a smaller subset then a Lasso model may be preferable.
- We could remove less important variables.
- We can do that manually by examining p-values of coefficients and discarding those variables whose coefficients are not significant.
- But this can become tedious for classification problems with many independent variables
Lasso Regression
- Originally introduced in geophysics literature in 1986
- The least absolute shrinkage and selection operator (Lasso) model was rediscovered and popularized in 1996 by Robert Tibshirani
- It is an alternative to Ridge regression that has a small modification to the penalty in the objective function.
-
Rather than the $L_2$ penalty we use the following $L_1$ penalty $\lambda\sum_{j=1}^p \beta_j $ in the objective function.
Lasso penalty pushes coefficients to zero
{ heigth=60% }
Lasso improves the model with regularization and conducts automated feature selection.
The reduction of coefficients
- 15 variables for $\text{log}(\lambda)=-5$
- 12 variables for $\text{log}(\lambda)=-1$
- 3 variables for $\text{log}(\lambda)=1$
When a data set has many features, Lasso can be used to identify and extract those features with the largest (and most consistent) signal.
Implementation Lasso regression to ames data
- Implementing Lasso follows the same logic as implementing the Ridge model, we just need to switch $\alpha = 1$ within
glmnet.
ames_lasso<-glmnet(x=ames_train_x,y=ames_train_y,alpha=1)
A quick drop in number of features
- Very large coefficients for ols (highly correlated)
- As model is constrained - these noisy features are pushed to 0.
- CV is necessary to determine right value for $\lambda$
plot(ames_lasso, xvar = "lambda")

Tuning with cv.glmnet
cv.glmnetwithalpha=1is used to perform cv.
ames_lasso<-cv.glmnet(x=ames_train_x,y=ames_train_y,alpha=1)
names(ames_lasso)
## [1] "lambda" "cvm" "cvsd" "cvup" "cvlo"
## [6] "nzero" "name" "glmnet.fit" "lambda.min" "lambda.1se"
MSE for cross validation
- MSE can be minimized with $-6\leq log (\lambda) \leq -4$
- Also the number of features can be reduced ($156 \leq p \leq 58$)
plot(ames_lasso)

Minimum and one standard error MSE and $\lambda$ values.
min(ames_lasso$cvm) # minimum MSE
## [1] 0.02246344
ames_lasso$lambda.min # lambda for this min MSE
## [1] 0.00332281
# 1 st.error of min MSE
ames_lasso$cvm[ames_lasso$lambda == ames_lasso$lambda.1se]
## [1] 0.02482119
ames_lasso$lambda.1se # lambda for this MSE
## [1] 0.01472211
MSE within one standard error
- The advantage of identifying the $\lambda$ with an MSE within one standard error becomes more obvious.
- If we use the $\lambda$ that drives the minimum MSE we can reduce our feature set from 307 down to less than 160.
- There is some variability with this MSE and we can assume that we can achieve a similar MSE with a slightly more constrained model (only 63 features).
- If describing and interpreting the predictors is an important outcome of your analysis, this will help.
Model with minimum MSE
plot(ames_lasso, xvar = "lambda")
abline(v=log(ames_lasso$lambda.min),col="red",lty="dashed")
abline(v=log(ames_lasso$lambda.1se),col="red",lty="dashed")

Lasso for other models than least squares
-
Though originally defined for least squares, Lasso regularization is easily extended to a wide variety of statistical models including generalized linear models, generalized estimating equations, proportional hazards models, and M-estimators, in a straightforward fashion.
-
Lasso’s ability to perform subset selection relies on the form of the constraint and has a variety of interpretations including in terms of geometry, Bayesian statistics, and convex analysis.
-
The Lasso is closely related to basis pursuit denoising.
Advantages and Disadvantages
- Similar to Ridge, the Lasso pushes many of the collinear features towards each other rather than allowing for one to be wildly positive and the other wildly negative.
- Unlike Ridge, the Lasso will actually push coefficients to zero and perform feature selection.
- This simplifies and automates the process of identifying those feature most influential to predictive accuracy.
Rcode for plotting influential variables
coef(ames_lasso, s = "lambda.1se") %>%
tidy() %>%
filter(row != "(Intercept)") %>%
ggplot(aes(value, reorder(row, value), color = value > 0)) +
geom_point(show.legend = FALSE) +
ggtitle("Influential variables") +
xlab("Coefficient") +
ylab(NULL)
Plot Influential variables
MSE for Ridge and Lasso
# minimum Ridge MSE
min(ames_ridge$cvm)
## [1] 0.01955871
# minimum Lasso MSE
min(ames_lasso$cvm)
## [1] 0.02246344
Elastic net
-
Elastic-Net is a compromise between Lasso and Ridge.
-
Elastic-Net penalizes a mix of both absolute and squared size.
- The ratio of the two penalty types should be tuned.
- The overall strength should also be tuned.
Elastic Nets
A generalization of the Ridge and Lasso models is the Elastic Net (Zou and Hastie, 2005), which combines the two penalties.
Summary overview
- A result of Lasso is that typically when two strongly correlated features are pushed towards zero, one may be pushed fully to zero while the other remains in the model.
- The process of one being in and one being out is not very systematic.
- In contrast, the Ridge regression penalty is a little more effective in systematically reducing correlated features together.
- The advantage of the Elastic Net model is that it enables effective regularization via the Ridge penalty with the feature selection characteristics of the Lasso penalty.
Implementation
alpha=.5performs an equal combination of penalties
lasso <- glmnet(ames_train_x, ames_train_y, alpha = 1.0)
elastic1 <- glmnet(ames_train_x, ames_train_y, alpha = 0.25)
elastic2 <- glmnet(ames_train_x, ames_train_y, alpha = 0.75)
ridge <- glmnet(ames_train_x, ames_train_y, alpha = 0.0)
The four model results plottet
Tuning the Elastic Net model
- $\lambda$ is the primary tuning parameter in Ridge and Lasso models.
- With elastic nets, we want to tune the $\lambda$ and the $\alpha$ parameters.
- To set up our tuning, we create a common
fold_id, which just allows us to apply the same CV folds to each model.
# maintain the same folds across all models
fold_id <- sample(1:10, size = length(ames_train_y),
replace=TRUE)
Creation of a tuning grid
- We then create a tuning grid that searches across a range of alphas from 0-1, and empty columns where we’ll dump our model results into.
# search across a range of alphas
tuning_grid <- tibble::tibble(
alpha = seq(0, 1, by = .1),
mse_min = NA,
mse_1se = NA,
lambda_min = NA,
lambda_1se = NA
)
Iteration over $\alpha$ values - Elastic Net
Now we can iterate over each $\alpha$ value, apply a CV Elastic Net, and extract the minimum and one standard error MSE values and their respective $\lambda$ values.
for(i in seq_along(tuning_grid$alpha)) {
# fit CV model for each alpha value
fit <- cv.glmnet(ames_train_x, ames_train_y,
alpha = tuning_grid$alpha[i],
foldid = fold_id)
# extract MSE and lambda values
tuning_grid$mse_min[i]<-fit$cvm[fit$lambda==fit$lambda.min]
tuning_grid$mse_1se[i]<-fit$cvm[fit$lambda==fit$lambda.1se]
tuning_grid$lambda_min[i]<-fit$lambda.min
tuning_grid$lambda_1se[i]<-fit$lambda.1se
}
The resulting tuning grid
tuning_grid
## # A tibble: 11 x 5
## alpha mse_min mse_1se lambda_min lambda_1se
## <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0 0.0198 0.0227 0.141 0.623
## 2 0.1 0.0205 0.0237 0.0365 0.134
## 3 0.2 0.0210 0.0243 0.0182 0.0736
## 4 0.3 0.0213 0.0249 0.0122 0.0539
## 5 0.4 0.0215 0.0249 0.00912 0.0404
## 6 0.5 0.0216 0.0250 0.00729 0.0323
## 7 0.6 0.0217 0.0255 0.00608 0.0296
## 8 0.7 0.0218 0.0255 0.00521 0.0253
## 9 0.8 0.0219 0.0255 0.00456 0.0222
## 10 0.9 0.0219 0.0255 0.00405 0.0197
## 11 1 0.0220 0.0256 0.00365 0.0177
Plot the MSE
- If we plot the MSE $\pm$ one standard error for the optimal $\lambda$ value for each alpha setting, we see that they all fall within the same level of accuracy.
- We could select a full Lasso model with $\lambda=0.02062776$, gain the benefits of its feature selection capability and reasonably assume no loss in accuracy.
tuning_grid %>%
mutate(se = mse_1se - mse_min) %>%
ggplot(aes(alpha, mse_min)) +
geom_line(size = 2) +
geom_ribbon(aes(ymax = mse_min + se, ymin = mse_min - se),
alpha = .25) +
ggtitle("MSE +/- one standard error")
MSE +/- one standard error

Predicting
- With the preferred model, you can
predictthe same model on a new data set. - The only caveat is you need to supply predict an s parameter with the preferred models $\lambda$ value.
- E.g., here we create a Lasso model, with a minimum MSE of 0.022.
# some best model
cv_lasso <- cv.glmnet(ames_train_x, ames_train_y, alpha = 1.0)
min(cv_lasso$cvm)
## [1] 0.02036225
- I use the minimum $\lambda$ value to predict on the unseen test set and obtain a slightly lower MSE of 0.015.
# predict
pred <- predict(cv_lasso, s = cv_lasso$lambda.min, ames_test_x)
mean((ames_test_y - pred)^2)
## [1] 0.02040651
The package caret - Classification and Regression Training
**Vignette for the caret package **
library(caret)
train_control <- trainControl(method = "cv", number = 10)
caret_mod <- train(x = ames_train_x,y = ames_train_y,
method = "glmnet",
preProc = c("center", "scale", "zv", "nzv"),
trControl = train_control,
tuneLength = 10)
Output for caret model
caret_mod
## glmnet
##
## 2054 samples
## 307 predictor
##
## Pre-processing: centered (113), scaled (113), remove (194)
## Resampling: Cross-Validated (10 fold)
## Summary of sample sizes: 1849, 1848, 1847, 1850, 1849, 1849, ...
## Resampling results across tuning parameters:
##
## alpha lambda RMSE Rsquared MAE
## 0.1 0.0001335259 0.1529759 0.8626587 0.09899496
## 0.1 0.0003084622 0.1529697 0.8626702 0.09899085
## 0.1 0.0007125878 0.1527312 0.8630945 0.09882473
## 0.1 0.0016461703 0.1523040 0.8638461 0.09858775
## 0.1 0.0038028670 0.1518043 0.8647353 0.09829651
## 0.1 0.0087851163 0.1511589 0.8658996 0.09792048
## 0.1 0.0202947586 0.1512482 0.8658861 0.09846301
## 0.1 0.0468835259 0.1542133 0.8616162 0.10095388
## 0.1 0.1083070283 0.1613716 0.8525304 0.10562397
## 0.1 0.2502032890 0.1783894 0.8378278 0.11798124
## 0.2 0.0001335259 0.1530144 0.8625906 0.09901019
## 0.2 0.0003084622 0.1529054 0.8627844 0.09893138
## 0.2 0.0007125878 0.1526093 0.8633042 0.09873037
## 0.2 0.0016461703 0.1520597 0.8642754 0.09843475
## 0.2 0.0038028670 0.1515362 0.8652246 0.09801179
## 0.2 0.0087851163 0.1511130 0.8660407 0.09800962
## 0.2 0.0202947586 0.1531822 0.8629272 0.10015289
## 0.2 0.0468835259 0.1583264 0.8557192 0.10357302
## 0.2 0.1083070283 0.1713989 0.8406963 0.11319420
## 0.2 0.2502032890 0.2015887 0.8196545 0.13607965
## 0.3 0.0001335259 0.1530175 0.8625852 0.09901246
## 0.3 0.0003084622 0.1528417 0.8628943 0.09887464
## 0.3 0.0007125878 0.1524303 0.8636143 0.09862411
## 0.3 0.0016461703 0.1519261 0.8645281 0.09829674
## 0.3 0.0038028670 0.1513782 0.8655399 0.09792851
## 0.3 0.0087851163 0.1517901 0.8649707 0.09874489
## 0.3 0.0202947586 0.1548532 0.8604377 0.10124984
## 0.3 0.0468835259 0.1631931 0.8487612 0.10712019
## 0.3 0.1083070283 0.1804627 0.8317568 0.12000176
## 0.3 0.2502032890 0.2242488 0.8048855 0.15383093
## 0.4 0.0001335259 0.1530032 0.8626107 0.09900104
## 0.4 0.0003084622 0.1527835 0.8629949 0.09883071
## 0.4 0.0007125878 0.1522777 0.8638796 0.09853749
## 0.4 0.0016461703 0.1517960 0.8647653 0.09816823
## 0.4 0.0038028670 0.1512813 0.8657246 0.09790412
## 0.4 0.0087851163 0.1527729 0.8634153 0.09973034
## 0.4 0.0202947586 0.1567219 0.8575524 0.10244652
## 0.4 0.0468835259 0.1674084 0.8431901 0.11030906
## 0.4 0.1083070283 0.1900641 0.8220999 0.12713979
## 0.4 0.2502032890 0.2456477 0.7968665 0.17219318
## 0.5 0.0001335259 0.1529694 0.8626695 0.09897170
## 0.5 0.0003084622 0.1527206 0.8631031 0.09877948
## 0.5 0.0007125878 0.1521743 0.8640690 0.09847427
## 0.5 0.0016461703 0.1517015 0.8649410 0.09808645
## 0.5 0.0038028670 0.1514410 0.8654740 0.09816784
## 0.5 0.0087851163 0.1535069 0.8622717 0.10039370
## 0.5 0.0202947586 0.1585892 0.8546972 0.10366586
## 0.5 0.0468835259 0.1711919 0.8386196 0.11310210
## 0.5 0.1083070283 0.1988715 0.8144491 0.13394394
## 0.5 0.2502032890 0.2677720 0.7874492 0.19208881
## 0.6 0.0001335259 0.1529457 0.8627110 0.09894655
## 0.6 0.0003084622 0.1526476 0.8632329 0.09872871
## 0.6 0.0007125878 0.1521007 0.8642136 0.09841560
## 0.6 0.0016461703 0.1516403 0.8650668 0.09805262
## 0.6 0.0038028670 0.1516698 0.8651021 0.09848230
## 0.6 0.0087851163 0.1541771 0.8612517 0.10084110
## 0.6 0.0202947586 0.1606510 0.8515691 0.10511838
## 0.6 0.0468835259 0.1751425 0.8337697 0.11597593
## 0.6 0.1083070283 0.2072075 0.8085757 0.14014121
## 0.6 0.2502032890 0.2895134 0.7807597 0.21111363
## 0.7 0.0001335259 0.1529182 0.8627588 0.09892152
## 0.7 0.0003084622 0.1525582 0.8633865 0.09868078
## 0.7 0.0007125878 0.1520463 0.8643179 0.09835732
## 0.7 0.0016461703 0.1515912 0.8651754 0.09802856
## 0.7 0.0038028670 0.1519555 0.8646544 0.09881899
## 0.7 0.0087851163 0.1548004 0.8603025 0.10119692
## 0.7 0.0202947586 0.1627999 0.8483414 0.10680467
## 0.7 0.0468835259 0.1792009 0.8286142 0.11899322
## 0.7 0.1083070283 0.2158269 0.8020943 0.14685245
## 0.7 0.2502032890 0.3123587 0.7639857 0.23129960
## 0.8 0.0001335259 0.1528934 0.8628024 0.09889940
## 0.8 0.0003084622 0.1524796 0.8635221 0.09864034
## 0.8 0.0007125878 0.1519877 0.8644285 0.09830023
## 0.8 0.0016461703 0.1515406 0.8652735 0.09802800
## 0.8 0.0038028670 0.1522942 0.8641256 0.09918411
## 0.8 0.0087851163 0.1554853 0.8592392 0.10161487
## 0.8 0.0202947586 0.1649098 0.8451450 0.10842937
## 0.8 0.0468835259 0.1832344 0.8233507 0.12199727
## 0.8 0.1083070283 0.2242570 0.7962765 0.15379654
## 0.8 0.2502032890 0.3360986 0.7248864 0.25185135
## 0.9 0.0001335259 0.1528683 0.8628464 0.09887948
## 0.9 0.0003084622 0.1524011 0.8636585 0.09859931
## 0.9 0.0007125878 0.1519283 0.8645367 0.09825245
## 0.9 0.0016461703 0.1514541 0.8654270 0.09800839
## 0.9 0.0038028670 0.1526743 0.8635214 0.09958496
## 0.9 0.0087851163 0.1562250 0.8580869 0.10208126
## 0.9 0.0202947586 0.1667279 0.8425633 0.10988380
## 0.9 0.0468835259 0.1869510 0.8187765 0.12486550
## 0.9 0.1083070283 0.2328192 0.7897299 0.16118585
## 0.9 0.2502032890 0.3598416 0.6471565 0.27230177
## 1.0 0.0001335259 0.1528421 0.8628890 0.09885896
## 1.0 0.0003084622 0.1523370 0.8637698 0.09856552
## 1.0 0.0007125878 0.1518838 0.8646164 0.09822006
## 1.0 0.0016461703 0.1514135 0.8655058 0.09802221
## 1.0 0.0038028670 0.1530411 0.8629353 0.09994305
## 1.0 0.0087851163 0.1570340 0.8568153 0.10261590
## 1.0 0.0202947586 0.1683894 0.8402729 0.11115096
## 1.0 0.0468835259 0.1904441 0.8148559 0.12743546
## 1.0 0.1083070283 0.2409139 0.7855310 0.16832017
## 1.0 0.2502032890 0.3805446 0.5646772 0.29026265
##
## RMSE was used to select the optimal model using the smallest value.
## The final values used for the model were alpha = 0.2 and lambda
## = 0.008785116.
Which regularization method should we choose?
- There’s no “best” type of penalty. It depends on the dataset and the problem.
- We recommend trying different algorithms that use a range of penalty strengths as part of the tuning process
Advantages and Disadvantages
- The advantage of the Elastic Net model is that it enables effective regularization via the Ridge penalty with the feature selection characteristics of the Lasso penalty.
-
Elastic Nets allow us to control multicollinearity concerns, perform regression when $p>n$, and reduce excessive noise in our data so that we can isolate the most influential variables while balancing prediction accuracy.
- Elastic Nets, and regularization models in general, still assume linear relationships between the features and the target variable.
- We can incorporate non-additive models with interactions, but it is tedious and difficult for a large number of features.
- When non-linear relationships exist, its beneficial to start exploring non-linear regression approaches.
Further packages
# https://cran.rstudio.com/web/packages/biglasso/biglasso.pdf
install.packages("biglasso")
Resources and Links
- Myers (1994) Classical and Modern Regression with Applications
Links
A comprehensive beginners guide for Linear, Ridge and Lasso Regression